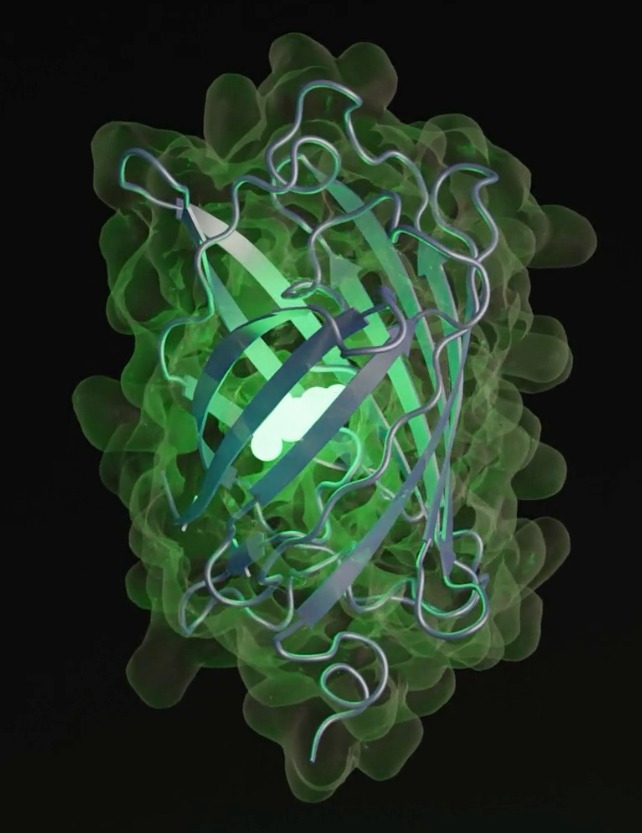
Synthesizing proteins—the fundamental building blocks of life—has long been a frontier of scientific discovery. Now, a groundbreaking AI model is pushing the boundaries of biology, generating entirely new proteins far beyond what nature has evolved.
Using EvolutionaryScale Model 3 (ESM3), scientists in the U.S. have successfully created esmGFP, a fluorescent green protein that shares just 58% of its structure with its closest natural counterpart, tagRFP. According to researchers, this is equivalent to simulating 500 million years of evolution—compressed into mere moments by AI.
AI-Powered Protein Design
ESM3 is a sophisticated AI model trained to design new proteins using an enormous dataset of evolutionary information. It analyzed:
- 3.15 billion protein sequences (the order of amino acids)
- 236 million protein structures (their 3D shapes)
- 539 million protein annotations (descriptive labels)
By identifying patterns in these vast datasets, ESM3 can predict and generate entirely new proteins with specific properties, similar to how language models like ChatGPT generate text based on vast amounts of written language.
“More than three billion years of evolution have encoded biology into the space of natural proteins,” writes the research team, led by Thomas Hayes, founder of EvolutionaryScale in New York.
“We show that language models trained at scale on evolutionary data can generate functional proteins that are far from any known natural proteins.”
A Functional Breakthrough in Protein Engineering
The real success of esmGFP isn’t just its novelty—it works. Like its natural relatives, it fluoresces, proving AI can generate viable, functional proteins. Fluorescent proteins are crucial in medical and biotech research, used as markers for tracking biological processes.
“We chose fluorescence because it is challenging to achieve, easy to measure, and one of the most beautiful mechanisms in nature,” the team explains.
Accelerating Evolution with AI
Biological evolution follows a slow, step-by-step process where each mutation must be viable. AI, however, allows scientists to bypass this trial-and-error approach, rapidly exploring new possibilities beyond natural evolution.
“Proteins exist in an interconnected space, where each is a neighbor to another through a single mutational step,” the researchers write.
“Evolution forms a network in this space, linking proteins through paths that nature can take over time.”
Unlike nature, AI isn’t restricted by evolutionary constraints—it can explore multiple alternative evolutionary pathways, generating proteins that may never have emerged through natural selection.
A Future of AI-Generated Proteins
Though AI-designed proteins still require validation and testing, the potential applications are staggering. From medicine to biomaterials, this technology could revolutionize how we design proteins for specific purposes—custom enzymes, advanced therapeutics, and novel materials may soon be just an AI prompt away.
“Protein language models do not explicitly follow evolution’s physical constraints,” the researchers explain, “but they can model the vast landscape of potential evolutionary paths.”
The findings have been published in Science, marking a significant leap in synthetic biology. As AI continues to evolve, so too may the very building blocks of life itself.